Hello.

Our lab focuses on the study of protein macromolecular complexes involved in epigenetic regulation of gene expression. Currently we study the methylcytosine binding domain 2 (MBD2) protein which recruits the Nucleosome Remodeling and Deacetylase (NuRD) complex to methylated DNA and silences expression of the associated gene.
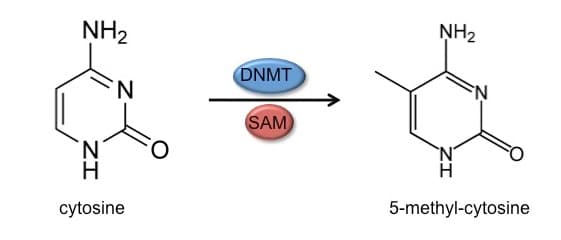
DNA methylation involves the addition of a methyl group to the C5 position of a cytosine base. This modification occurs in the context of CpG dinucleotides in double-stranded DNA in which the symmetrically related cytosines are methylated. This modification can be maintained across cell division since each daughter cell inherits one of the methylated strands.
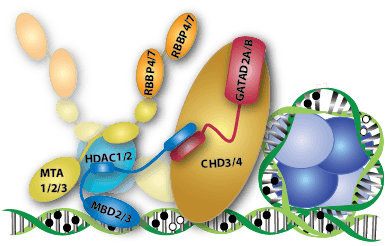
The NuRD complex comprises MBD2 or MBD3 and at least five additional proteins, each of which have multiple isoforms: HDAC1/2, RBBP4/7, MTA1/2/3, GATAD2A/B, and CHD3/4. We have been focusing our efforts on understanding how the MBD2/3 proteins bind and distribute on DNA and how they interact with other members of the complex.
A coiled-coil interaction between the GATAD2A and MBD2 proteins recruits the CHD3/4 nucleosome remodeler to the NuRD complex. We solved the structure of this interaction and have shown that a small peptide inhibitor blocks methylation dependent gene silencing. Based on this work, we are currently developing an inhibitor of complex formation to restore expression of methylated and silenced genes.
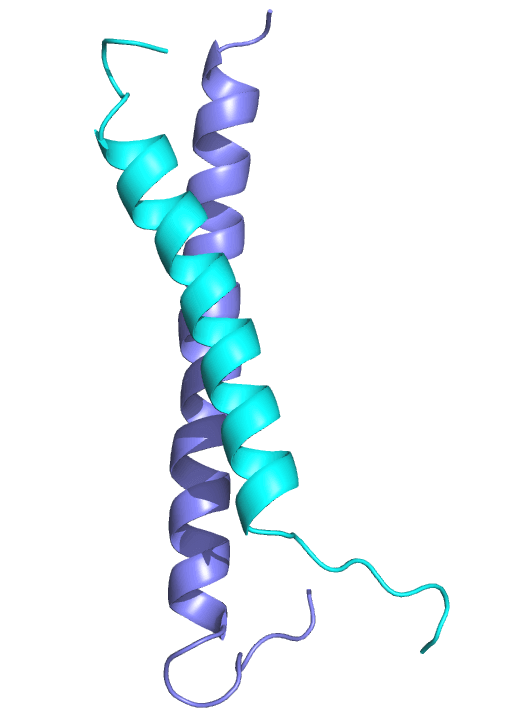
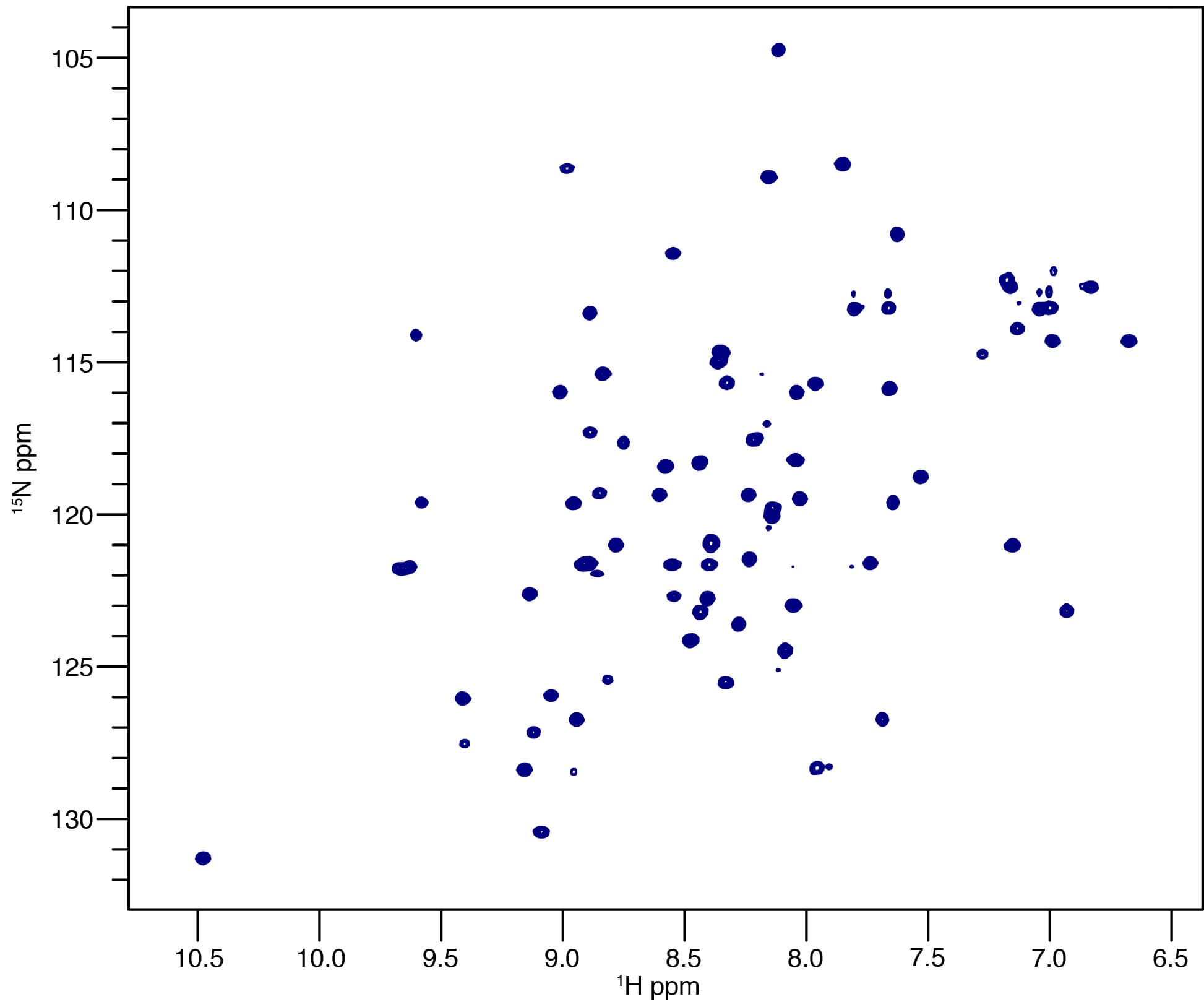
We use nuclear magnetic resonance (NMR) spectroscopy and other biophysical techniques including isothermal titration calorimetry, circular dichroism and surface plasmon resonance to investigate structure and dynamics of protein-protein and protein-DNA complexes.